About
Abstract:
|
Transmembrane proteins play crucial roles in various cellular processes. The biological effects of protein modifications on transmembrane proteins include phosphorylation for signal transduction and ion transport, acetylation for structure stability, attachment of fatty acids for membrane anchoring and association, as well as the glycosylation for substrates targeting, cell-cell interactions, and viruses infection. With the importance of PTMs functioning on transmembrane proteins, we are motivated to develop a database, topPTM, that integrates experimentally verified post-translational modifications (PTMs) from available databases and research articles, and annotates the PTM sites on transmembrane proteins with structural topology. The experimentally verified PTMs are mainly collected from public resources including dbPTM, Phospho.ELM, PhosphoSite, OGlycBase, and UbiProt. Additionally, due to the emerging evidences in nitric oxide (NO)-related pathway, the experimentally verified protein S-nitrosylation sites are manually extracted from approximately 200 S-nitrosylation-related research articles using a text mining approach. For transmembrane proteins, the information of membrane topologies is collected from TMPad, TOPDB, PDBTM, and OPM. In order to fully investigate the PTMs on transmembrane proteins, the UniProtKB protein entries containing the annotation of membrane protein and the information of membrane topology are regarded as potential transmembrane proteins. The structural topology of transmembrane proteins is represented by graphical visualization, as well as the PTMs. Moreover, the tertiary structure of PTM sites on transmembrane proteins is visualized by Jmol program. To delineate the structural correlation and consensus motif of these reported PTM sites, the topPTM database also provide structural analyses, including the membrane accessibility of PTM substrate sites, protein secondary and tertiary structures, protein domains, and cross-species conservations of each entry.
The topPTM is now freely accessible via http://topPTM.cse.yzu.edu.tw. The database is regularly updated upon collecting new data from continuously surveying research articles and public resources. |
Flowchart:
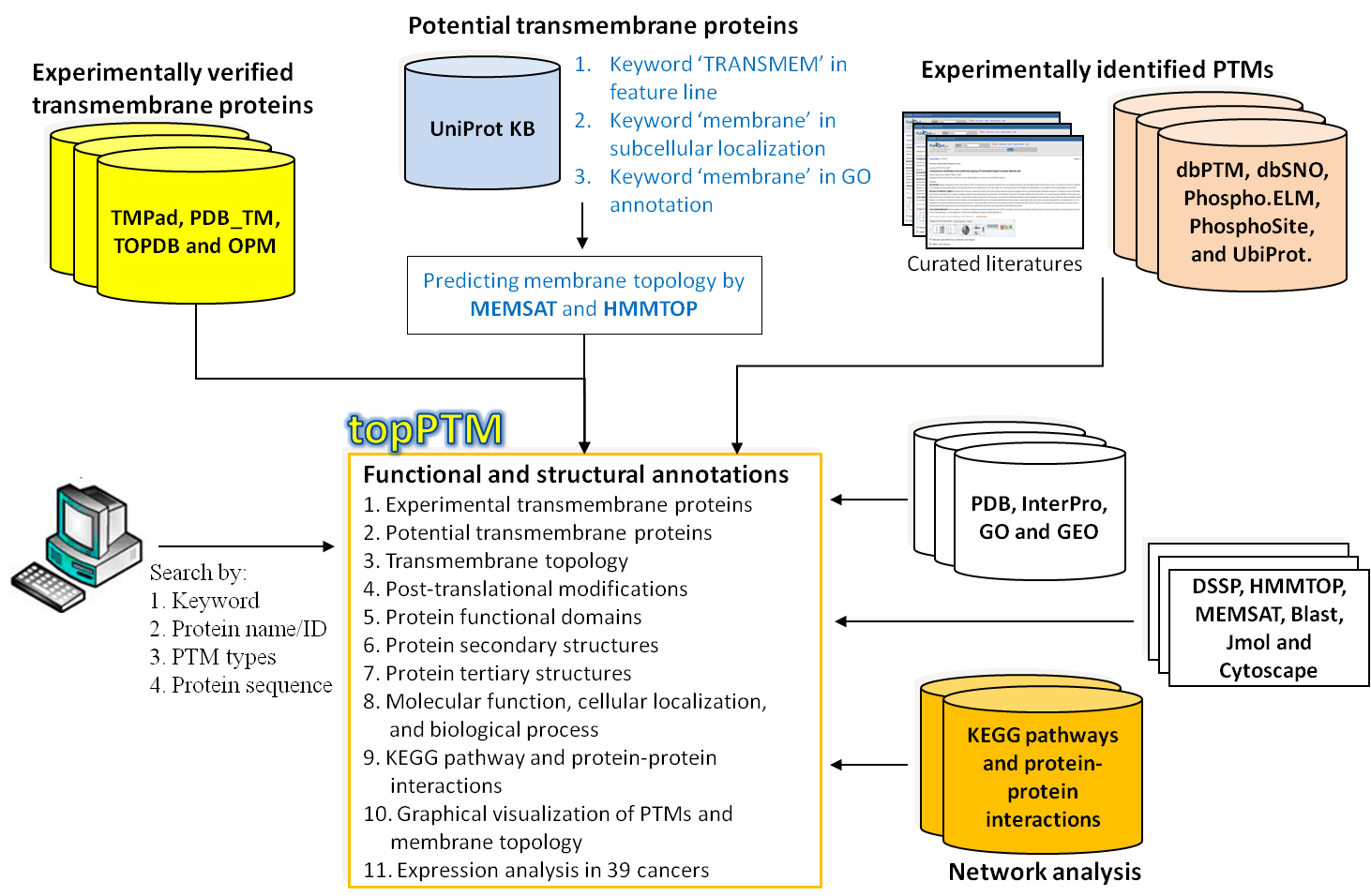
●Figure. The system flow of constructing topPTM database.
Statistics:
| Resource | Number of Experimentally Verified TM Protein (at least one transmembrane domain are Experimental) |
Number of Potential TM Protein | Number of Experimentally Verified with α (Protein with at least one transmembrane helical domain) |
Number of Potential with α | Number of Experimentally Verified with β (Protein with at least one transmembrane beta stranded domain) |
Number of Potential with β |
| TMPad | 379 | - | 379 | - | 0 | - |
| OPM | 651 | - | 1435 | - | 44 | - |
| TOPDB | 1479 | - | 667 | - | 91 | - |
| PDBTM | 785 | - | 556 | - | 96 | - |
UniProtKB |
590 |
73444 | 260 | 72343 | 13 |
972 |
Total |
2234 |
73444 | 2067 | 72603 | 170 |
985 |
| Transmembrane in Special Cell membrane | Protein Count Number |
| Virion | 1022 |
| Nuclear | 94 |
| Mitochondrial | 1014 |
| Chloroplast | 44 |
| Exoplasmic loop | 3 |
| Forespore | 3 |
| Lumenal | 4509 |
| Intragranular | 12 |
| Intravacuolar | 2 |
| Vacuolar | 204 |
| Vesicular | 245 |
| Periplasmic(Bacteria) | 3743 |
| Peroxisome | 51 |
| PTM Instance Type | Number of PTM sites on TM proteins (experimental) | Number of PTM sites on TM proteins (poteintial) |
| Phosphoserine | 4489 | 7009 |
| Glycosylation N-linked (GlcNAc...) | 2550 | 63197 |
| Phosphothreonine | 1072 | 2078 |
| Phosphotyrosine | 900 | 2675 |
| Glycosylation O-linked (GalNAc...) | 233 | 844 |
| N6-acetyllysine | 182 | 510 |
| S-palmitoyl cysteine | 136 | 2341 |
| N6-(retinylidene)lysine | 75 | 170 |
| N-acetylalanine | 63 | 149 |
| N-acetylmethionine | 42 | 129 |
| N-formylmethionine | 31 | 113 |
| Glutamate methyl ester (Glu) | 29 | 33 |
| Sulfotyrosine | 24 | 334 |
| N-acetylserine | 19 | 78 |
| Pyrrolidone carboxylic acid | 19 | 33 |
| Nitrated tyrosine | 19 | 14 |
| N-myristoyl glycine | 17 | 142 |
| Glutamate methyl ester (Gln) | 16 | 15 |
| Glycosylation O-linked (Xyl...) | 14 | 166 |
| N-acetylthreonine | 13 | 382 |
| 4-aspartylphosphate | 11 | 525 |
| Phosphohistidine | 10 | 367 |
| Glycosylation C-linked (Man) | 10 | 15 |
| Omega-N-methylated arginine | 9 | 25 |
| Glycosylation N-linked (Glc...) | 6 | 16 |
| Blocked amino end (Met) | 6 | 0 |
| Deamidated asparagine | 6 | 0 |
| N2-acetylarginine | 5 | 48 |
| Deamidated glutamine | 5 | 1 |
| Leucine amide | 5 | 1 |
| Glycosylation O-linked (Hex...) | 5 | 0 |
| Glycosylation O-linked (GlcNAc...) | 4 | 25 |
| N6-malonyllysine | 4 | 20 |
| N6-palmitoyl lysine | 4 | 0 |
| N6-myristoyl lysine | 3 | 51 |
| S-8alpha-FAD cysteine | 3 | 15 |
| Methylhistidine | 3 | 14 |
| Lysine amide | 3 | 6 |
| ADP-ribosylarginine | 3 | 5 |
| Glutamine amide | 3 | 5 |
| N6,N6-dimethyllysine | 3 | 5 |
| S-farnesyl cysteine | 3 | 5 |
| Dimethylated arginine | 3 | 4 |
| Omega-N-methylarginine | 3 | 4 |
| (3S)-3-hydroxyasparagine | 3 | 3 |
| S-nitrosocysteine | 3 | 3 |
| Blocked amino end (Gln) | 3 | 0 |
| S-diacylglycerol cysteine | 2 | 169 |
| N-palmitoyl cysteine | 2 | 168 |
| FMN phosphoryl threonine | 2 | 51 |
| 4-hydroxyproline | 2 | 20 |
| S-stearoyl cysteine | 2 | 8 |
| O-(5'-phospho-RNA)-serine | 2 | 6 |
| N-acetylglycine | 2 | 3 |
| Glycosylation N-linked (GalNAc...) | 2 | 2 |
| Cysteine methyl ester | 2 | 2 |
| Hydroxyproline | 2 | 0 |
| Phenylalanine amide | 2 | 0 |
| Sulfoserine | 2 | 0 |
| Aspartyl aldehyde | 1 | 103 |
| (3R)-3-hydroxyasparagine | 1 | 9 |
| GPI-anchor amidated serine | 1 | 7 |
| N6-methyllysine | 1 | 6 |
| Glycosylation O-linked (Man6P...) | 1 | 4 |
| 2',4',5'-topaquinone | 1 | 4 |
| 3-oxoalanine (Cys) | 1 | 4 |
| N-acetylglutamate | 1 | 3 |
| O-palmitoyl threonine | 1 | 3 |
| Glycosylation O-linked (GlcNAc) | 1 | 2 |
| ADP-ribosylcysteine | 1 | 2 |
| GPI-anchor amidated aspartate | 1 | 2 |
| N-acetylvaline | 1 | 1 |
| Asymmetric dimethylarginine | 1 | 0 |
| Blocked amino end (Ala) | 1 | 0 |
| Glutamic acid 1-amide | 1 | 0 |
| N-acetylcysteine | 1 | 0 |
| N-formylglycine | 1 | 0 |
| S-glutathionyl cysteine | 1 | 0 |
| 4-carboxyglutamate | 0 | 43 |
| N6-(pyridoxal phosphate)lysine | 0 | 24 |
| Glycosylation O-linked (Gal...) | 0 | 16 |
| O-(pantetheine 4'-phosphoryl)serine | 0 | 12 |
| O-palmitoyl serine | 0 | 8 |
| Pentaglycyl murein peptidoglycan amidated threonine | 0 | 7 |
| (3R)-3-hydroxyaspartate | 0 | 5 |
| N-methylphenylalanine | 0 | 5 |
| Pros-8alpha-FAD histidine | 0 | 5 |
| GPI-anchor amidated asparagine | 0 | 5 |
| GPI-anchor amidated alanine | 0 | 2 |
| Glycosylation O-linked (Ara...) | 0 | 1 |
| N6-carboxylysine | 0 | 1 |
| N-methylmethionine | 0 | 1 |
| O-(5'-phospho-RNA)-tyrosine | 0 | 1 |
| Valine amide | 0 | 1 |
| GPI-like-anchor amidated serine | 0 | 1 |
Total |
10115 |
82292 |
| PTM Instance Type | Experimented PTM site on ... | Non-Experimented PTM site on ... | ||||||||||
| Extracellular | Cytoplasmic | Transmembrane | Extracellular | Cytoplasmic | Transmembrane | |||||||
Exp |
Non-Exp |
Exp |
Non-Exp |
Exp |
Non-Exp |
Exp |
Non-Exp |
Exp |
Non-Exp |
Exp |
Non-Exp |
|
| Phosphoserine | 2 | 189 | 62 | 2523 | 3 | 25 | 0 | 204 | 104 | 4990 | 4 | 46 |
| Glycosylation N-linked (GlcNAc...) | 80 | 1898 | 0 | 0 | 0 | 0 | 367 | 43134 | 0 | 0 | 0 | 6 |
| Phosphothreonine | 1 | 78 | 16 | 560 | 0 | 14 | 3 | 93 | 78 | 994 | 2 | 60 |
| Phosphotyrosine | 2 | 56 | 13 | 673 | 2 | 27 | 3 | 97 | 11 | 2246 | 2 | 68 |
| Glycosylation O-linked (GalNAc...) | 40 | 123 | 0 | 0 | 0 | 0 | 6 | 697 | 0 | 0 | 0 | 0 |
| N6-acetyllysine | 0 | 1 | 3 | 38 | 6 | 17 | 0 | 1 | 4 | 109 | 1 | 75 |
| S-palmitoyl cysteine | 0 | 0 | 11 | 82 | 0 | 29 | 0 | 8 | 98 | 1784 | 1 | 208 |
| N6-(retinylidene)lysine | 0 | 0 | 0 | 0 | 3 | 72 | 0 | 1 | 0 | 0 | 1 | 168 |
| N-acetylalanine | 0 | 1 | 0 | 12 | 0 | 0 | 0 | 2 | 0 | 38 | 0 | 0 |
| N-acetylmethionine | 1 | 0 | 2 | 18 | 0 | 0 | 21 | 0 | 0 | 35 | 0 | 0 |
| N-formylmethionine | 0 | 0 | 1 | 8 | 0 | 2 | 0 | 0 | 0 | 11 | 0 | 2 |
| Glutamate methyl ester (Glu) | 0 | 0 | 0 | 16 | 0 | 0 | 0 | 0 | 0 | 28 | 0 | 0 |
| Sulfotyrosine | 3 | 20 | 0 | 1 | 0 | 0 | 11 | 292 | 0 | 2 | 0 | 0 |
| N-acetylserine | 0 | 0 | 2 | 7 | 0 | 0 | 0 | 0 | 0 | 58 | 0 | 0 |
| Pyrrolidone carboxylic acid | 2 | 12 | 0 | 0 | 0 | 0 | 0 | 33 | 0 | 0 | 0 | 0 |
| Nitrated tyrosine | 0 | 2 | 0 | 6 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 4 |
| N-myristoyl glycine | 0 | 1 | 0 | 3 | 0 | 0 | 0 | 26 | 0 | 14 | 0 | 0 |
| Glutamate methyl ester (Gln) | 0 | 3 | 0 | 14 | 0 | 0 | 0 | 0 | 0 | 11 | 0 | 0 |
| Glycosylation O-linked (Xyl...) | 1 | 13 | 0 | 0 | 0 | 0 | 0 | 163 | 0 | 0 | 0 | 0 |
| N-acetylthreonine | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 4-aspartylphosphate | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 22 | 0 | 0 |
| Phosphohistidine | 0 | 0 | 0 | 6 | 0 | 0 | 0 | 1 | 0 | 152 | 0 | 0 |
| Glycosylation C-linked (Man) | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 7 | 0 | 0 | 0 | 0 |
