
| Transmembrane (TM) proteins play crucial roles in various cellular processes. The biological effects of post-translational modifications (PTMs) on TM proteins include phosphorylation for signal transduction and ion transport, acetylation for structure stability, attachment of fatty acids for membrane anchoring and association, as well as glycosylation for cell-cell communication. With the importance of PTMs functioning on TM proteins, we are motivated to develop a public resource, topPTM, which is a new module of dbPTM for identifying the functional PTM sites on TM proteins with structural topology. The experimentally verified information of transmembrane topologies was integrated from TMPad, TOPDB, PDBTM and OPM. In addition to the PTMs obtained from dbPTM, we manually extracted the experimentally verified PTM sites from research articles by text mining. In an attempt to provide a full investigation of PTM sites on TM proteins, all the UniProtKB protein entries containing the annotations of membrane localization and transmembrane topology were regarded as potential TM proteins. Then, two effective tools were utilized to annotate the structural topology of the potential TM proteins. The transmembrane topology of TM proteins is represented by graphical visualization, as well as the PTM sites. To delineate the structural correlation between the PTM sites and transmembrane topologies, the tertiary structure of PTM sites on TM proteins is visualized by the Jmol program. The topPTM also provides functional investigation of PTMs, network analysis involving metabolic pathways and protein-protein interactions, and expression profiles in cancers for TM proteins. The topPTM is now freely accessible via http://topPTM.cse.yzu.edu.tw. The database is regularly updated upon collecting new data from continuously surveying research articles and public resources. |
| Flowchart: |
| The system flowchart of constructing topPTM is presented in Figure 1. Experimentally verified TM proteins annotated with membrane topology information are mainly collected from PDB_TM (14), OPM (17), TOPDB (18), and TMPad (20). After the removal of redundant protein entries, a total of 2234 TM proteins containing experimentally curated annotations of transmembrane topology remained. Additionally, a candidate set of TM proteins is extracted from UniProtKB by choosing protein entries which contain the keyword “TRANSMEM” in feature (“FT”) line, the localization of “membrane”, and the information of transmembrane topology. The candidate TM proteins are further filtered using HMMTOP (26) and MEMSAT (27) to determine their transmembrane topologies. |
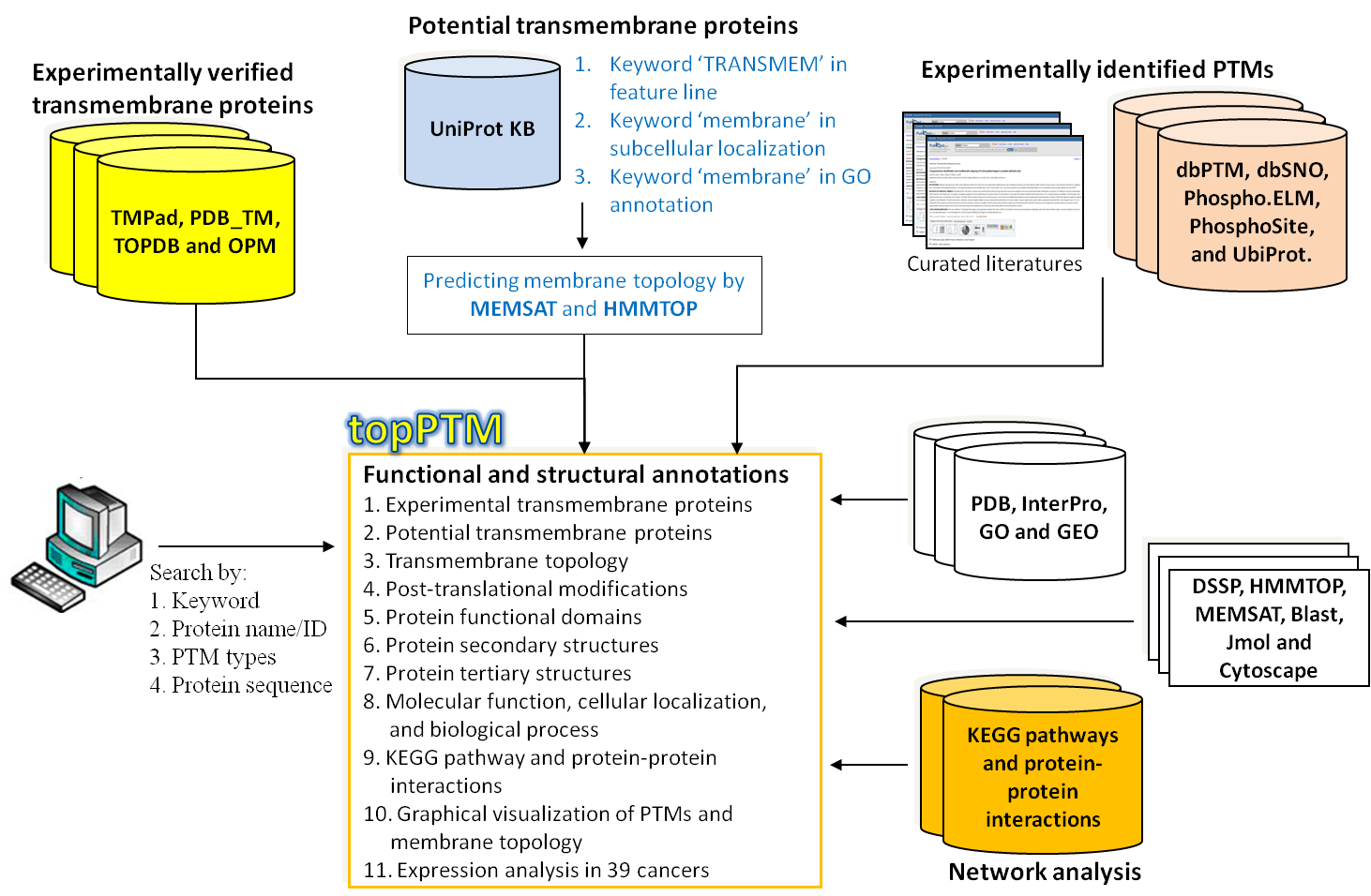
●Figure 1. The system flow of constructing topPTM database.
